GP v/s Deep GP on 2d data#
Author: Nipun Batra, Zeel B Patel
In this notebook, we will be comparing three models (all RBF kernel) on a simulated 2d data:
GP
GP with ARD
Deep GP
import GPy
import numpy as np
import pyDOE2
from sklearn.metrics import mean_squared_error
# Plotting tools
from matplotlib import pyplot as plt
from matplotlib import rc
rc('font', size=16)
rc('text', usetex=True)
import warnings
warnings.filterwarnings('ignore')
# GPy: Gaussian processes library
import GPy
We generate pseudo data using the following function,
np.random.seed(0)
X = pyDOE2.doe_lhs.lhs(2, 9, random_state=0)
func = lambda x: ((x[:, 0]+2)*(2-x[:, 1])**2)/3
y = func(X)
plt.scatter(X[:,0], X[:, 1],s=y*50);
GP without ARD#
k_2d = GPy.kern.RBF(input_dim=2, lengthscale=1)
m = GPy.models.GPRegression(X, y.reshape(-1, 1), k_2d)
m.optimize();
m
Model: GP regression
Objective: -5.479737333080651
Number of Parameters: 3
Number of Optimization Parameters: 3
Updates: True
| GP_regression. | value | constraints | priors |
|---|---|---|---|
| rbf.variance | 72.28075242747792 | +ve | |
| rbf.lengthscale | 3.382677032484262 | +ve | |
| Gaussian_noise.variance | 9.922601887760885e-06 | +ve |
x_1 = np.linspace(0, 1, 40)
x_2 = np.linspace(0, 1, 40)
X1, X2 = np.meshgrid(x_1, x_2)
X_new = np.array([(x1, x2) for x1, x2 in zip(X1.ravel(), X2.ravel())])
Y_true = func(X_new)
Y_pred, Y_cov = m.predict(X_new)
Y_95 = 2*np.sqrt(Y_cov)
fig, ax = plt.subplots(1,2,figsize=(12,4))
mp = ax[0].contourf(X1, X2, Y_pred.reshape(*X1.shape), levels=30)
fig.colorbar(mp, ax=ax[0])
ax[0].set_title(f'Predictive mean, RMSE = {mean_squared_error(Y_true, Y_pred, squared=False).round(7)}');
mp = ax[1].contourf(X1, X2, Y_95.reshape(*X1.shape), levels=30)
ax[1].set_title(f'Predictive variance (95\% confidence)')
fig.colorbar(mp, ax=ax[1]);
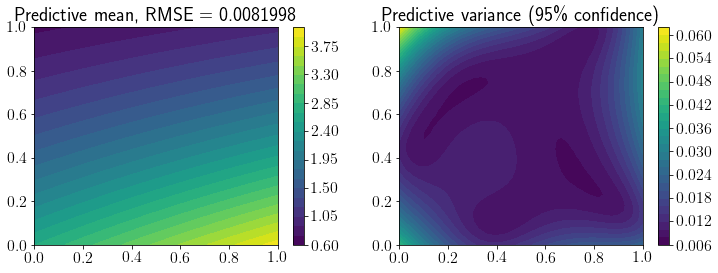
GP with ARD#
k_2d_ARD = GPy.kern.RBF(input_dim=2, lengthscale=1, ARD=True)
m = GPy.models.GPRegression(X, y.reshape(-1, 1), k_2d_ARD)
m.optimize();
m
Model: GP regression
Objective: -6.917350273214053
Number of Parameters: 4
Number of Optimization Parameters: 4
Updates: True
| GP_regression. | value | constraints | priors |
|---|---|---|---|
| rbf.variance | 40.53729208947549 | +ve | |
| rbf.lengthscale | (2,) | +ve | |
| Gaussian_noise.variance | 4.3169473160951275e-29 | +ve |
Y_pred, Y_cov = m.predict(X_new)
Y_95 = 2*np.sqrt(Y_cov)
fig, ax = plt.subplots(1,2,figsize=(12,4))
mp = ax[0].contourf(X1, X2, Y_pred.reshape(*X1.shape), levels=30)
fig.colorbar(mp, ax=ax[0])
ax[0].set_title(f'Predictive mean, RMSE = {mean_squared_error(Y_true, Y_pred, squared=False).round(7)}');
mp = ax[1].contourf(X1, X2, Y_95.reshape(*X1.shape), levels=30)
ax[1].set_title(f'Predictive variance (95\% confidence)')
fig.colorbar(mp, ax=ax[1]);
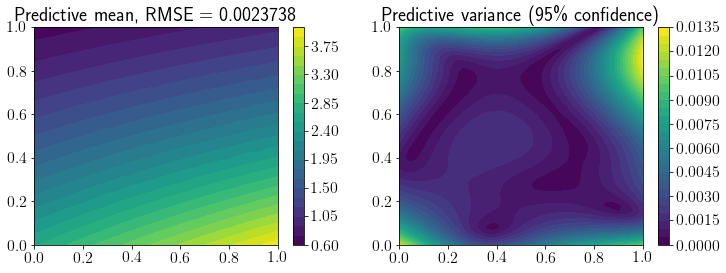
m.parameters[0].lengthscale
| index | GP_regression.rbf.lengthscale | constraints | priors |
|---|---|---|---|
| [0] | 5.80242511 | +ve | |
| [1] | 1.80657554 | +ve |
| deepgp. | value | constraints | priors |
|---|---|---|---|
| obslayer.inducing inputs | (4, 1) | ||
| obslayer.rbf.variance | 23.808937388338997 | +ve | |
| obslayer.rbf.lengthscale | 4.451925378737478 | +ve | |
| obslayer.Gaussian_noise.variance | 0.0020011072507906376 | +ve | |
| obslayer.Kuu_var | (4,) | +ve | |
| obslayer.latent space.mean | (9, 1) | ||
| obslayer.latent space.variance | (9, 1) | +ve | |
| layer_1.inducing inputs | (4, 2) | ||
| layer_1.rbf.variance | 30.67508979761088 | +ve | |
| layer_1.rbf.lengthscale | (2,) | +ve | |
| layer_1.Gaussian_noise.variance | 0.0012460891277525125 | +ve | |
| layer_1.Kuu_var | (4,) | +ve |
Y_pred, Y_cov = m.predict(X_new)
Y_95 = 2*np.sqrt(Y_cov)
fig, ax = plt.subplots(1,2,figsize=(12,4))
mp = ax[0].contourf(X1, X2, Y_pred.reshape(*X1.shape), levels=30)
fig.colorbar(mp, ax=ax[0])
ax[0].set_title(f'Predictive mean, RMSE = {mean_squared_error(Y_true, Y_pred, squared=False).round(3)}');
mp = ax[1].contourf(X1, X2, Y_95.reshape(*X1.shape), levels=30)
ax[1].set_title(f'Predictive variance (95\% confidence)')
fig.colorbar(mp, ax=ax[1]);
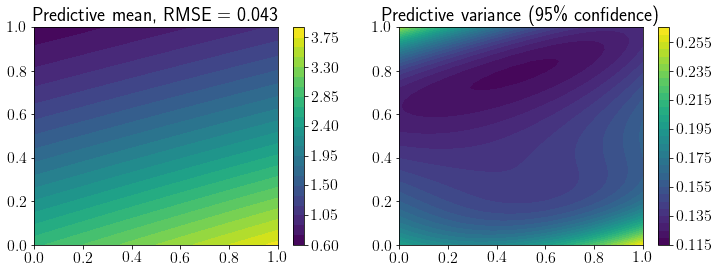
Not working Pinball_plot : lawrennd/talks

