Automatic relevance determination (ARD)#
Author: Zeel B Patel
import scipy.stats
import GPy
from scipy.optimize import minimize
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import rc
import warnings
warnings.filterwarnings('ignore')
%matplotlib inline
rc('text', usetex=True)
rc('font', size=16)
To understand the concept of ARD, let us generate a symthetic dataset where all features are not equally important.
np.random.seed(0)
N = 400
X = np.empty((N, 3))
y = np.empty((N, 1))
cov = [[1,0,0,0.99],[0,1,0,0.6],[0,0,1,0.1],[0.99,0.6,0.1,1]]
samples = np.random.multivariate_normal(np.zeros(4), cov, size=N)
X[:,:] = samples[:,:3]
y[:,:] = samples[:,3:4]
print('Correlation between X1 and y', np.corrcoef(X[:,0], y.ravel())[1,0])
print('Correlation between X2 and y', np.corrcoef(X[:,1], y.ravel())[1,0])
print('Correlation between X3 and y', np.corrcoef(X[:,2], y.ravel())[1,0])
Correlation between X1 and y 0.7424364387053712
Correlation between X2 and y 0.4771760788020134
Correlation between X3 and y 0.07463999808005005
Let us fit a GP model with a common lengthscale for all features.
model = GPy.models.GPRegression(X, y, GPy.kern.RBF(input_dim=3, ARD=False))
model.optimize()
model
Model: GP regression
Objective: 346.85990977337315
Number of Parameters: 3
Number of Optimization Parameters: 3
Updates: True
| GP_regression. | value | constraints | priors |
|---|---|---|---|
| rbf.variance | 111.47290536431103 | +ve | |
| rbf.lengthscale | 26.862865061479035 | +ve | |
| Gaussian_noise.variance | 0.3083839240873474 | +ve |
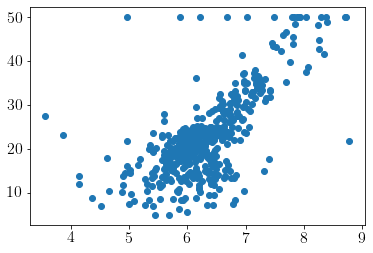
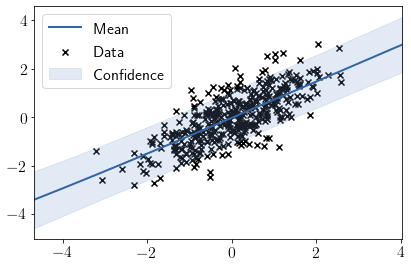
Visualizing fit over \(X_1\)
model.plot(visible_dims=[0]);
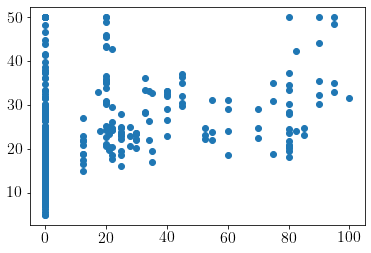
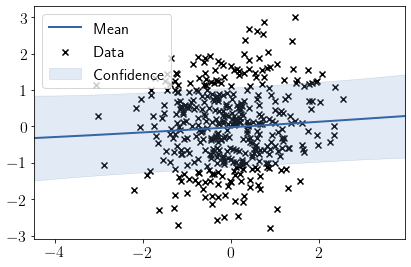
Visualizing fit over \(X_3\)
model.plot(visible_dims=[2]);
Now, let us turn on the ARD and see the values of lengthscales learnt.
model = GPy.models.GPRegression(X, y, GPy.kern.RBF(input_dim=3, ARD=True))
model.optimize()
model.kern.lengthscale
| index | GP_regression.rbf.lengthscale | constraints | priors |
|---|---|---|---|
| [0] | 22.24437029 | +ve | |
| [1] | 33.16175836 | +ve | |
| [2] | 143.39745522 | +ve |
| index | GP_regression.rbf.lengthscale | constraints | priors |
|---|---|---|---|
| [0] | 288.47316598 | +ve | |
| [1] | 1958.66785662 | +ve | |
| [2] | 96.63008923 | +ve | |
| [3] | 522.58028671 | +ve | |
| [4] | 262.09440505 | +ve | |
| [5] | 3.15337665 | +ve | |
| [6] | 267.59752767 | +ve | |
| [7] | 2.07260971 | +ve | |
| [8] | 154.25307337 | +ve | |
| [9] | 218.28124322 | +ve | |
| [10] | 48.57585913 | +ve | |
| [11] | 282.84461914 | +ve | |
| [12] | 22.50407090 | +ve |